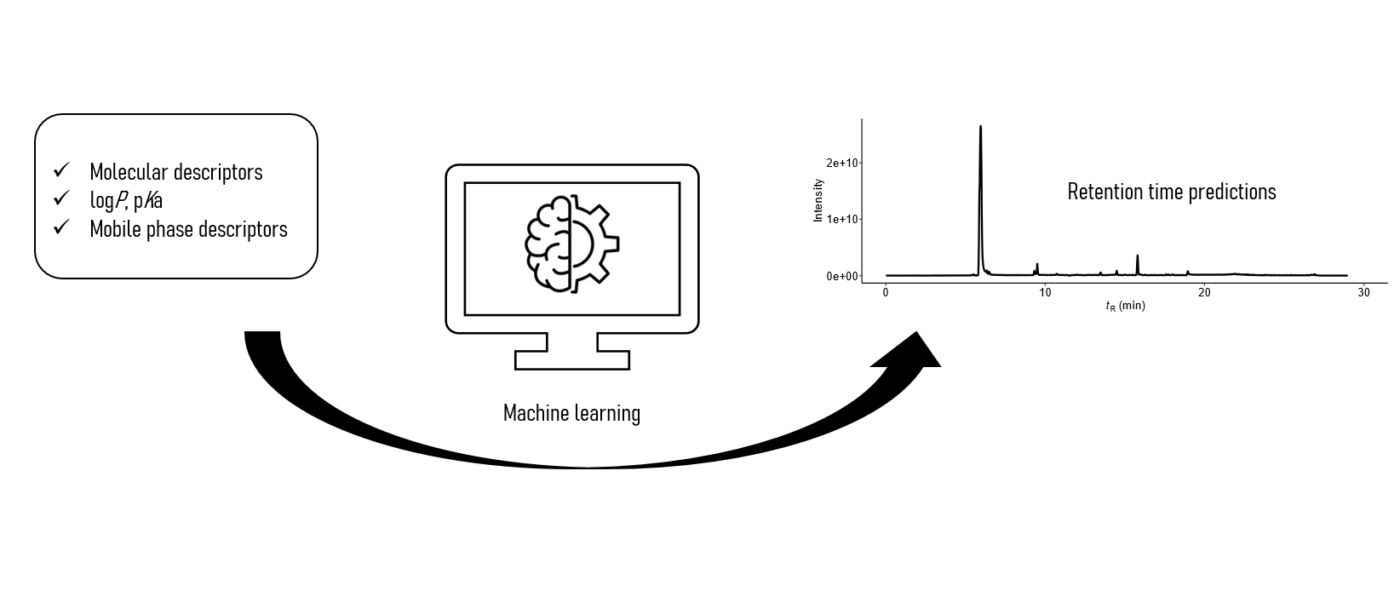
Retention time is a crucial information for structural elucidation of unknown chemicals in non-targeted analysis using liquid chromatography coupled to high resolution mass spectrometry via electrospray ionization (LC/ESI/HRMS). Most of the predictive models of retention time are developed in limited chromatographic conditions, mostly for reversed phase and HILIC mode and in acidic conditions. However, retention time may change significantly depending on the chromatographic conditions used. In our recent paper, we developed and validated MultiConditionRT to predict retention times in 4 different chromatographic mechanisms, and 20 different mobile phases.
MultiConditionRT was developed based on 78 representative compounds, selected from the NORMAN compounds in MassBank (S1). Different chromatographic systems involved C18 reversed phase, mixed-mode, biphenyl and HILIC. As organic solvents both acetonitrile and methanol were used, while water phase contained one of seven additives (formic acid, acetic acid, TFA, ammonium formate, ammonium acetate, ammonium bicarbonate, ammonia) in a pH range from 2.1 to 10.0.
The root mean square error (RMSE) of MultiConditionRT was 1.55 min for C18 reversed phase, 1.79 min for mixed mode, 1.93 min for HILIC and 1.56 min for biphenyl. The predictions of MultiConditionRT were validated internally using an in-house dataset containing 151 compounds with an RMSE of 2.32 min. Using a projection approach, MultiConditionRT was used for predicting retention times on an external dataset of 356 compounds. The external validation of MultiConditionRT yielded an RMSE of 2.68 min. If you are interested in applying MultiConditionRT on your chromatographic systems you can find all of the machine learning models from GitHub. Feel free to contact us for more information.
In the future we aim to implement MultiConditionRT in the workflow of non-targeted analysis to aid the annotation of features together with library spectra matching, isotope pattern and adduct formation.