Recently we have published a paper on advancement in predicting ionization efficiency in ESI and using this for estimating the concentration of pesticides in cereal samples without the need for analytical standards.
In this paper, we explored the possibility to use machine learning for fast and accurate prediction of ionization efficiency. Previously we have carefully analysed the factors affecting the ionization of different compounds in ESI positive and negative mode. It is clear that both the compound structure and eluent compositions are essential for accurately predicting ionization efficiency. In the current paper, we combined data about 353 compounds in ESI+ and 106 compounds in ESI- for more than 106 eluent combinations. The dataset incorporates compounds with logP from -4 to +8 and eluents from 100% water phase to 100% organic modifier and with pH from 2.1 to 10.7 for both ESI positive and negative mode. Altogether 3139 and 1286 ionization efficiency values were collected for this dataset. Notably, the data were collected on several instruments in the labs of collaboration partners around the world (Claude Bernard Lyon 1, Janssen R&D in Belgium, University of Tartu, Technical University of Denmark, University of North Carolina Greensboro).
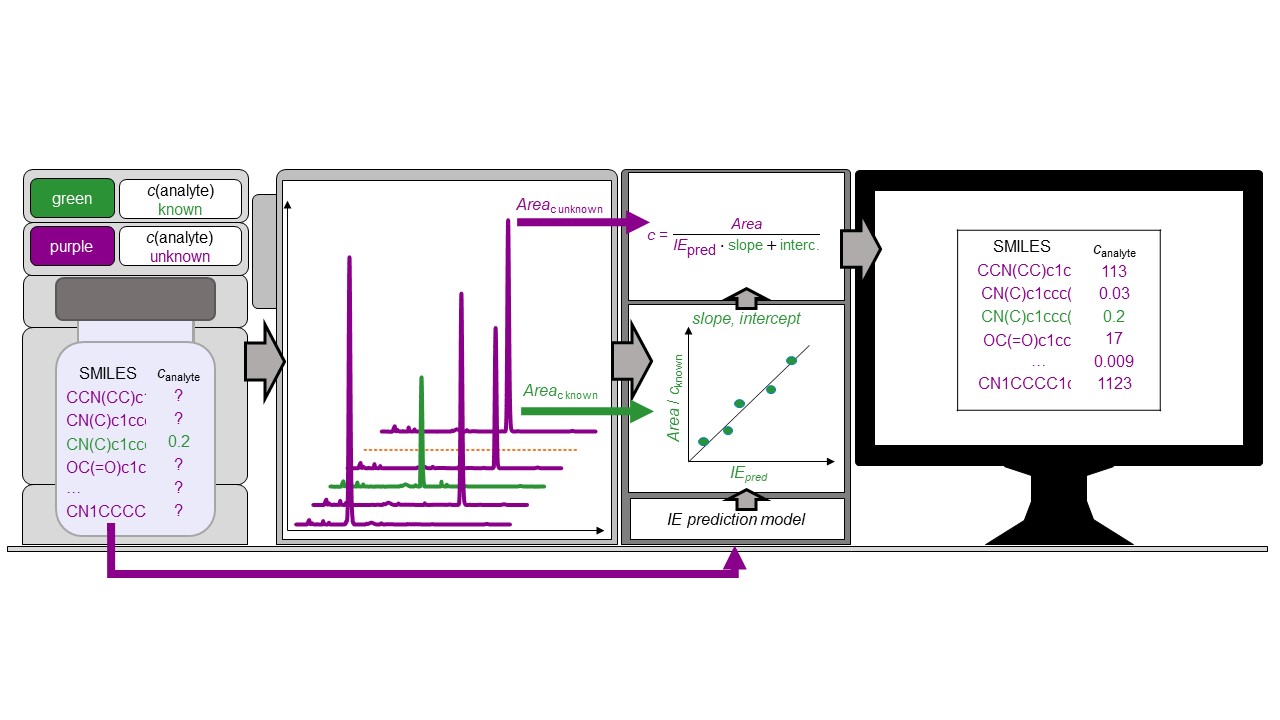
The ionization efficiencies are predicted based on the PaDEL fingerprint descriptors and eluent parameters. Different machine learning algorithms were tested and optimized. The best prediction accuracy was obtained with random forest regression. The ionization efficiency values ranged over more than a factor of 100 million but the machine learning algorithm allowed a mean prediction error better than a factor of 3 for the test set.
The predicted ionization efficiencies were thereafter used to estimate the concentration of pesticides in cereals. The extracts of cereals had been fortified with 35 pesticides at the concentration range from 3.6 nM to 0.35 mM. The average concentration prediction error was 5.4x. In food control, the concentration of the contaminants is combined with the predicted toxic endpoints. The uncertainty in the toxic endpoints is around an order of magnitude. Based on the rules of combining uncertainty, we can say that the prevailing source of uncertainty is the toxic endpoint prediction and obtained concentration prediction accuracy is well compatible for this purpose. We are sure that the ability to quantify the compounds tentatively identified with non-targeted analysis makes combining non-targeted analysis with targeted analysis even more compelling. This concentration prediction approach is also made available for everyone through Quantem.
All in all, combining fingerprint descriptors and random forest allows predicting the concentrations of compounds detected with non-targeted LC/HRMS rapidly and accurately. We are preparing a number of quantitative non-targeted analysis for environmental and food analysis and hope to show the results to you very soon. Stay tuned!