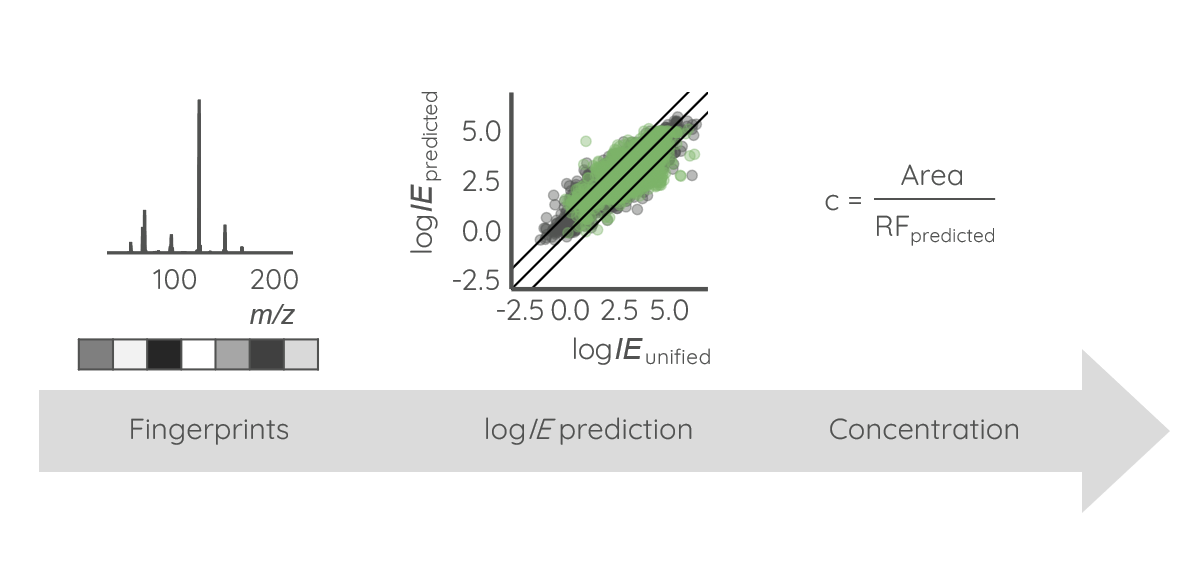
Non-targeted screening is helping us to discover the complex chemical space of environmental samples. Due to the large number of detected chemicals, prioritization is necessary to direct focus on chemicals posing a risk to biota and humans. Commonly, key information for risk assessment is the exposure to the respective chemical. Unfortunately, the detected signal intensity is poorly correlated to the concentration across different chemicals, see Figure 1. That is because some chemicals ionize better than others due to their physicochemical properties and, therefore, a matching standard is required for precise concentration estimation of a chemical.
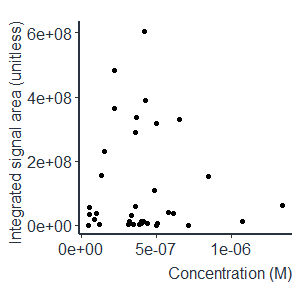
Figure 1. Higher signal intensity does not necessarily mean a higher concentration of a chemical. In the example of NORMAN interlaboratory comparison calibrants, the maximum concentration difference is 28× while the maximum fold difference in signal intensities is 10,900×.
While there are a number of alternatives to using analytical standards for quantification (such as using a surrogate standard for quantification that is structurally similar or close in eluting time, or logIE prediction), majority of these methods require that the structure of the chemical is known prior to estimating its concentration. In a model MS2Quant trained here, we are harvesting structural information from the fragmentation spectra. For this, SIRIUS+CSI:FingerID is used to predict the probability of the presence or absence of structural fingerprints which are used as input for ionization efficiency prediction.
MS2Quant was trained using ionization efficiency data for 1191 unique chemicals that span 8 orders of magnitude. Modelling resulted in root mean square errors (RMSE) of 0.55 (3.5×) and 0.80 (6.3×) log-units for the training and test set, respectively. To estimate the concentration of detected LC/HRMS features prior to identification, the following steps need to be taken:
- Measure a set of calibrants within the same run with your sample. These chemicals should cover a wide range of ionization efficiency values and are used to convert the predicted ionization efficiency value of the suspects into measurement-specific response factors.
- Predict the probability of the presence/absence of structural fingerprints for your suspects. For this, SIRIUS+CSI:FingerID can be used which needs the fragmentation spectra of suspects as input.
- Install MS2Quant and use it for quantification. The function needs calibrants and suspects information, gradient program and path to SIRIUS calculation results as input.
MS2Quant is available as an R-package on our GitHub page together with guidelines for installation, testing and application. Additionally, we would like to thank Rick Helmus who is integrating MS2Quant and MS2Tox into patRoon workflow for easy implementation in the NTS workflow.