Non-targeted screening with liquid chromatography-electrospray high-resolution mass spectrometry (LC/ESI/HRMS) is revealing hundred to thousands of contaminants in the water. We have recently proposed a way to quantify these contaminants based on the estimated LC/ESI/HRMS ionization efficiencies. But how to know how well such tools are performing? Well, we need to compare with classical methods that are based on analytical standards. Very recently we had a rare chance to compare different quantification methods back-to-back based on a unique set of 341 contaminants quantified with targeted LC/HRMS methods in 31 groundwater samples from Switzerland by Juliane Hollender group at EAWAG. The work is now available from Anal Bioanal Chem.
Methods to quantify suspects without analytical standards
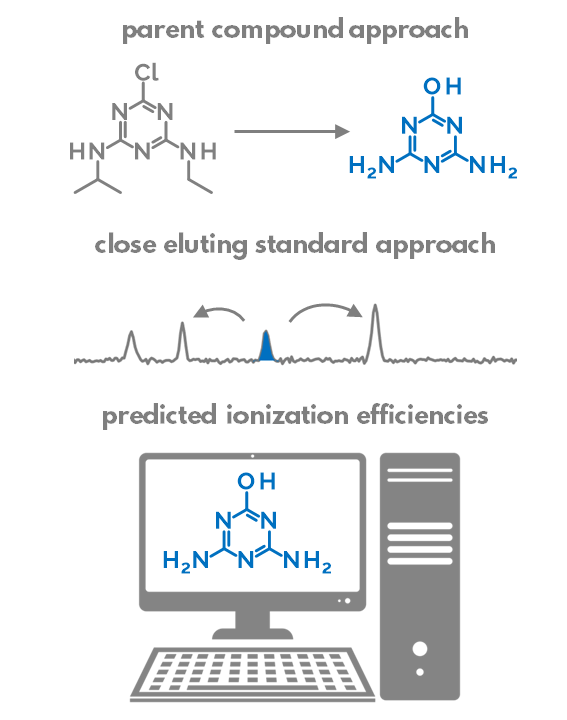
We compared the ability of (1) paren compound approach, (2) close eluting standard approach, and (3)to quantify contaminants detected from surface water samples with non-targeted LC/ESI/HRMS.
The parent compound approach uses the calibration graph of a parent compound to quantify the transformation products. The assumption is that the transformation products (TPs) are sufficiently similar to the parent compound and would, therefore, ionize very similarly in the parent compound in the ESI. We found that this assumption is appropriate if the structural changes from parent compound to TP are small. Most importantly, the hydrophobicity (logP) and acid-base properties should not change significantly during transformation. Sometimes, however, the changes are substantial: e.g. when full alkyl chains are cleaved off, the TPs become much more hydrophobic in comparison to parent compounds and their ionization efficiencies drop dramatically. In some cases, the transformation may even result in the switching of the detection polarity from ESI positive to negative or vice versa.
So which compounds are similar to each other and can be used for quantification? Pieke et al. have put forth the hypothesis that the internal standards eluting close to the suspects could be sufficiently similar in their ionization efficiency and, therefore, used for quantification of the suspects. Indeed, the retention time in reversed-phase chromatography is strongly correlated with the logP of the compound. Similarly, the compounds with higher logP tend to have higher ionization efficiencies, but only if the acid-base properties of these compounds are similar.
Lastly, we can also try to predict the ionization efficiency of each compound in electrospray based on the structure of this compound and the LC conditions. We have developed a random forest based machine-learning algorithm to predict the ionization efficiencies for different compounds. The model uses PaDEL descriptors to describe the compound structure and it also accounts for mobile phase composition: organic modifier percent at the retention time of the suspect, pH, organic modifier type, and buffer type. We have previously also applied this strategy for suspect quantification of pesticides in food. Though this approach can be applied to all compounds, the nature of using molecular fingerprints makes the predictions be applicable to compounds that are sufficiently similar to the training set compounds. E.g. compounds with completely new functionality will not be accurately predicted.
The performance of analytical standard free quantification
The best accuracy was observed with the predicted ionization efficiency-based quantification. The mean error of concentration prediction for the groundwater samples was less than 2x, and all of the 74 micropollutants detected in the groundwater were quantified with an error below 10x. The runner-up was the closest eluting standard approach with a mean prediction error of slightly over 3x and the lowest performance was observed for quantification with the parent compounds with a mean error of slightly below 4x.
The software tool, both web-based and desktop, for ionization efficiency-based quantification is being developed by Quantem Analytics.